Samuel Garrard and Dr. Kenneth Christensen, Department of Chemistry and Biochemistry
Introduction:
Pathological angiogenesis, or new blood vessel formation, is required for the progression of many diseases, including cancer, macular degeneration, and many other pathologies. Effective inhibition of angiogenesis would greatly augment current treatments for both cancer and eye disease. Previous research has implicated CMG2 as a key regulator of angiogenesis in vivo. Despite this, the normal function of CMG2 in the human body is incompletely understood.
A better understanding of CMG2 function is necessary to developing anti-angiogenic treatments targeting CMG2. This can be achieved by identifying natural ligands of CMG2 in vivo and measuring binding affinity of CMG2-ligand interactions. Knowledge of these interactions will connect CMG2 with known cellular pathways, and aid in any future development of anti-angiogenic drugs targeting CMG2. It is known that CMG2 binds to extracellular matrix proteins such as collagen IV, collagen VI, fibronectin, and laminin; therefore, I chose to measure affinity of CMG2 for these proteins, as well as for collagen I, which binds the CMG2 homolog tumor endothelial marker 8 protein (TEM8).
Methodology:
I adapted the traditional ELISA assay to create a physiologically relevant method for measuring CMG2-ligand affinity. A GST–CMG2–AviTag construct (which I developed and expressed previously) was used as the primary antibody, and 96-well polyethylene plates were coated with matrix proteins as antigen. This setup was chosen because it best mimicked the environment of cell-matrix interactions. Streptavidin–HRP was used as the secondary antibody, and tetramethylbenzidine (TMB) was used for detection. To determine affinity of CMG2 for each matrix protein, I added a range of GST–CMG2–AviTag concentrations, from 2 μM to 20 nM, to each plate. I then used the resulting absorbance data to generate binding isotherms and calculate a KD for each interaction.
Controls:
Bacillus anthracis protective antigen (PA), whose binding to CMG2 is well characterized, was coated onto the plate as a positive control. Wells coated only with blocking buffer (5% BSA in HBS w/ 2mM Mg2+, Ca2+) served as a negative control. An additional negative control, which included removal of Mg2+ and Ca2+ from all buffers and addition of EDTA to the CMG2 conjugate solution, was also used. However, this was an ineffective control since CMG2-matrix interactions were not disrupted by removal of metal ions from solution (see discussion for further insights).
Results:
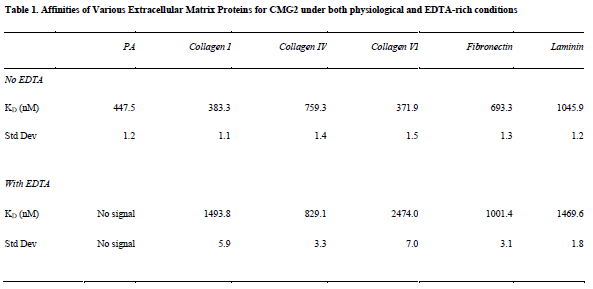
I successfully calculated KD values for each matrix protein tested, as well as PA (see Table 1). KD values in the presence of EDTA were also calculated for all proteins except for PA, which generated no signal.
Discussion:
My data indicates that all matrix proteins tested bind CMG2 with high specificity, and with similar affinities (370 nM to 1.5 μM). These affinities are similar to those of interactions between matrix proteins and integrins, which are structurally similar to CMG2. My values are therefore not surprising, although I did not expect all matrix proteins to bind CMG2 with similar affinity. This indicates that CMG2 may bind a consensus motif present in each of these matrix proteins.
Additionally, I found that matrix interactions are not inhibited by removal of metal cations from solution and from CMG2 via EDTA. This came as a a surprise, since the CMG2–PA interaction requires a metal ion-dependent adhesion site domain (MIDAS). However, other hydrophobic proteins (BSA and casein) were unable both to produce the signal intensity and pattern of the CMG2-matrix interactions, indicating that these interactions are indeed specific. This novel discovery indicates that CMG2 has multiple binding sites, each of which may perform a separate function.
Conclusion:
I have characterized several CMG2–matrix interactions, and, for the first time, generated relative affinities for several matrix ligands of CMG2. These indicate that CMG2 does not preferentially bind any of these matrix proteins. This ‘promiscuous’ binding may indicate a consensus binding motif for CMG2-matrix interactions, which would mirror the mechanism of many known matrix-receptor interactions. Additionally, the understanding that CMG2–matrix interactions are not MIDAS-mediated may explain why CMG2 can mediate both pro- and anti-angiogenic signaling, as is suggested by data from our collaborator, Michael Rogers, at Harvard University.
It should be noted that due to the time constraints associated with this project, these results are only preliminary, and have only partially been replicated. Any conclusions are therefore preliminary and require validation.