Russell, Robert C
Development of a Directed Forward Genetic Screen Using CRISPR Libraries
Jonathon Hill, Physiology and Developmental Biology
Introduction
Thousands of genes control the development of our bodies. These genes must be
activated at the proper place and time in order for development to proceed correctly.
The technique that we are developing will improve our ability to identify such genes,
thus providing insights to the mechanisms underlying development processes. One in
33 live births in the United States is impacted by a birth defect. Of these, congenital
heart defects (CHDs) are the most common, affecting nearly 1% of births in the United
States.1 For this reason, we are developing a technique that will identify key contributors
in heart development.
Forward genetic screens are a common and effective way to identify genes underlying
developmental defects. These screens are done by randomly mutation animal genes
and then breeding those animals to identify phenotypes. The gene causing the desired
phenotype is then genetically mapped. Currently, this is most commonly done using Nethyl-
N-nitrosourea (ENU) or another mutagenic chemical. ENU has two major
limitations: 1) it is a dangerous mutagen for lab personnel to handle and 2) it causes
mutations anywhere on the genome. This second limitation requires a large number of
families to be generated because many mutations will be in noncoding regions or genes
not involved in the process of interest. These problems can be solved by extracting
RNA from a specific tissue and timepoint to create a CRISPR guide RNA library.
Generating guide RNAs from a specific time/place will focus mutations to the coding
regions of relevant genes, making forward genetic screens more efficient.
Methodology
We started developing this technique by generating a normalized cDNA library using
whole zebrafish embryo to maximize the amount of cDNA available. RNA was first
extracted using the Trizol Mini Kit (Zymo Research). A double-stranded cDNA library
was then constructed using the SMART cDNA Library Construction Kit (Clontech) and
normalized using the Trimmer 2 cDNA Normalization Kit (Evrogen). The normalized
library was analyzed by qPCR to test for relative expression of a panel of reference
genes. Once we confirmed that we could make a normalized cDNA library from whole
zebrafish embryo, we repeated the process using RNA that was extracted from
zebrafish hearts 48 hours post fertilization. After normalization of the heart specific
library we proceeded with the creation of the CRISPR gRNA library. To do this we used
a restriction enzyme that cut at a protospacer adjacent motif sequence (PAM site).
Once the PAM site was cut, an MmeI adapter was attached. MmeI restriction enzyme
cut 20 bp 5’ to the original PAM site and was then removed. A T7 promoter was added
to the 5’ end and an sgRNA region is added to the 3’ end, which will be used by the
Cas9 protein to hold the gRNA in place.
Results
Thus far we have created our CRISPR gRNA library from RNA extracted from the heart.
A qPCR before and after normalization shows that our normalization procedure works
well (See Figure 1). After the last step in attaching the guide RNA regions we ran the
product through a PAGE gel and cut out a band that was around 130 base pairs (See
Figure 2). This is the length of the gRNA sequences that will be used in the forward
genetic screens with CRISPR Cas9 mutations.
Discussion
Throughout the experiment there were several steps that took longer than what we had
initially expected. During the steps of ligating the proper promoters and gRNA
sequences to our target sequence, we followed a 29 step protocol used by Dr. Lane in
his publication Enzymatically Generated CRISPR Libraries for Genome Labeling and
Screening.2 To drastically reduce the number of steps required for this protocol we have
started another project that adds a restriction site into the sgRNAs and uses linkers
attached to magnetic beads.
While our CRISPR gRNA library is complete, there are further tests that must be
completed before injecting embryos and conducting forward genetic screens. Our next
step is to analyze the complexity of our library through high-throughput sequencing and
compare it to the zebrafish genome to ensure we have optimal target sites for
mutations.
Conclusion
Thus far, the objective data that we
have gained has lead us to believe that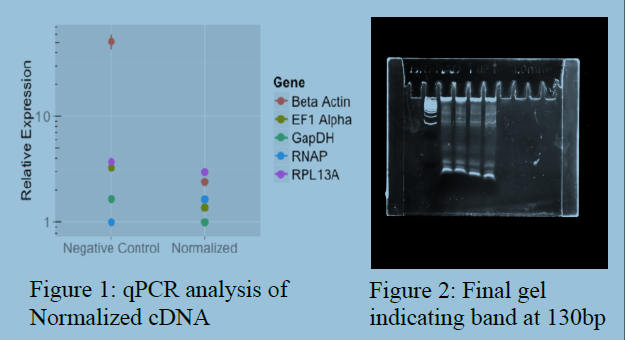
we have generated the appropriate
library. We had the appropriate DNA
concentrations at various steps and the
proper finishing length on our PAGE
gel. We will continue to analyze the
library and apply it in modified forward
genetic screens.
1 Center for Disease Control and Convention. (2015). Data and Statistics, Congenital Heart Defects. Retrieved from
http://www.cdc.gov/ncbddd/heartdefects/data.html#References on October 27, 2015
2 Lane AB, Strzelecka M, Ettinger A, Grenfell AW, Wittmann T, Heald R. (2015). Enzymatically Generated
CRISPR Libraries for Genome Labeling and Screening. Developmental Cell. 34(3):373-8.
