Pattison, Jenny Adele
The role of PAS kinase and Cbf1 in Cellular Metabolism
Faculty Mentor: Julianne H. Grose, Mircobiology and Molecular Biology
Introduction
The most recent National Institute of Health report concludes that 68.8% of American adults are considered to be overweight or obese1. With this trend in obesity, metabolic diseases such as heart disease, stroke, cancer, and diabetes are on the rise. The critical yet basic function of cellular homeostasis is a central component in controlling these diseases. Sensory protein kinases are essential in the phosphorylation of many protein substrates, allowing them to control several metabolic functions and appropriately allocate glucose, maintaining homeostasis in cells. PAS kinase is a sensory protein kinase that is highly conserved and plays a crucial role in cellular respiration. Although PAS kinase is a necessary component for essential metabolic functions, little is known about the molecular mechanisms behind these functions. We have recently discovered a key substrate of PAS kinase that affects glucose metabolism in the cell, Centromere binding factor 1 (Cbf1)2. Cbf1 regulates genes involved in respiration, and we have shown that the phosphorylation of Cbf1 by PAS kinase inhibits Cbf1, decreasing respiration in yeast cells. In agreement, when PAS kinase is knocked out of cells, there is a significant Cbf1-dependent increases in cellular respiration rates. Cbf1’s importance is further supplemented by its human homolog, USF1, which has been associated with hyperlipidemia in humans. We have recently shown that USF1, like its yeast homolog, rescues cellular respiration as well.
Methodology
Flow cytometry was utilized to quantify the total intracellular reactive oxygen species (cell-ROS) and mitochondrial total reactive oxygen species (mit-ROS) in knockout strains and overexpression strains of each identified respiratory regulators listed above. To measure cell-ROS, the oxidant sensitive, cell permeant fluorescent probe H2DCFDA was used. To measure mit-ROS, the oxidant-sensitivity, cell permeant fluorescent probe dihydrohodamine 123 (DHR123) was used. Strains were grown in 2% YPAD overnight and then diluted 1:50 in YPAraffinose and grown for four hours. Samples were aliquoted and immediately loaded with either H2DCFDA or DHR123 and incubated at 30’C for 2 hours in the dark. Cells were then diluted with PBS buffer and immediately quantified by flow cytometry. The fluorescence was monitored in the emission fluorescence channel FL1. The populations of cells were gated in the forward scatter and side scatter dot plots to eliminate dead cells and cellular debris. Flow-jo analytical software was used in order to identify novel differences in ROS levels between the strains.
Results
In order to find novel pathways in the regulation of respiration (both Cbf1-dependent and independent), I conducted a high copy suppressor screen for proteins that could repair the respiration defect of Cbf1-deficient yeast. I performed nine screens, plating over ten million transformants. Most of the suppressors retrieved were plasmids containing either full length or truncated Cbf1. However, I was able to identify seven novel targets responsible for respiration in Cbf1 deficient yeast: MCK1 (meiosis and centromere regulatory kinase), SFG1 (SuperFicial pseudohyphal growth), HMS1 (high-copy Mep suppressor), CCT4 (chaperonin containing TCP-1), NIT1 (NITrilase superfamily), COY1 (CASP of Yeast), IRT1 (IME1 Regulatory Transcript), PAL1 (protein of unkown function), RIM11 (regulator of IME2), and BLM10 (BLeoMycin resistance). Although each of these suppressors restores respiratory growth of our yeast, none of these suppressors has a previously described role in the regulation of respiration.
My second aim this summer was to identify changes in reactive oxygen species when these identified suppressors are not present in yeast. In normal cells, energy is produced primarily through mitochondrial oxidative phosphorylation. However, in high levels of glucose, cells will produce energy through high rates of glycolysis. This switch to glycolysis, known as the Warburg effect, is common in many proliferative cells3. Under some conditions, reactive oxygen species are given off in greater degrees when a cell switches to glycolytic growth.
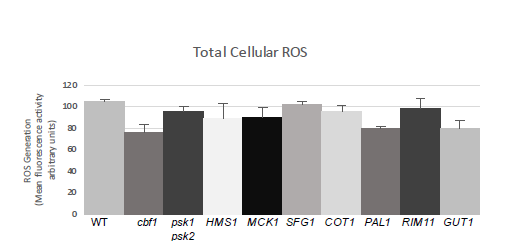
We tested the total cell-ROS and found that there was no significant difference between these three strains. We then tested the mit-ROS which revealed a significant difference between the wild-type and cbf1-deficient yeast as well as a significant difference between the psk1psk2-deficient yeast. We then tested the total cell-ROS and mit-ROS in HMS1-deficient, MCK1-deficient, SFG1-deficient, COT1-deficient, PAL1-deficient, RIM11-deficient, and GUT1-deficient strains and compared them to the three strains used in the previous experiment. Similar to the previous experiment, the mit-ROS assay did not show a significant difference between the strains. The cell-ROS showed decreases in both the PAL1-deficient yeast and GUT1-deficient yeast. It should be noted this assay was only performed once and as such I was not able to run an ANOVA on these samples to determine their statistical difference.
Discussion
Understanding how cells are regulating respiration is essential to understanding metabolic disease. As mentioned previously, the phosphorylation of Cbf1 by PAS kinase inhibits Cbf1 function, decreasing respiration in yeast and when PAS kinase is knocked out of cells, there is a significant Cbf1-dependent increase in cellular respiration rates. We therefore predicted and verified that there would be a noticeable difference in the reactive oxygen species produced between wild-type, cbf1-deficient, and psk1psk2-deficient yeast. This study also identifies novel regulation of reactive oxygen species by GUT1 and PAL1, as we see a novel decrease in reactive oxygen species level in both PAL1 and GUT1-deficient yeast. These results suggest these novel respiratory suppressors as excellent candidates to direct future respiratory studies towards.
Conclusion
Despite the increasing trend of metabolic disease, the mechanisms of PAS kinase and Cbf1 are largely unknown. By identifying suppressors of Cbf1 and understanding how they regulate cellular respiration we can increase our understanding of metabolic pathways as well as identify possible therapeutic targets for metabolic disease.
Sources
(1) “Overweight and Obesity Statistics.” Overweight and Obesity Statistics. National Institute of Diabetes and Digestive and Kidney Diseases, 1 Oct. 2012. Web. 14 Oct. 2015. <http://www.niddk.nih.gov/health-information/health-statistics/Pages/overweight-obesity-statistics.aspx>
(2) Demille, D., Bikman, B., Mathis, A., Prince, J., Mackay, J., Sowa, S., Hall, T., Grose, J., (2014). A Comprehensive Protein-protein Interactome for Yeast PAS Kinase 1 Reveals Direct Inhibition of Respiration Through the Phosphorylation of Cbf1. Molecular Biology of the Cell, 25(14), 2199-2215.
(3) Vander Heiden, M., Cantley, L., & Thompson, C. (2009). Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science, 324(5930), 1029-1033