King, Taylor
Damselflies of Patagonia: A phylogeographic study utilizing EPIC DNA and mitochondrial sequence markers
Faculty Mentor: Seth Bybee, Biology Department

Introduction
While working on my original project, “In Situ Hybridization of Opsins in Odonata,” I
encountered abundant setbacks that led to refocusing my efforts on a similar project. Patagonian
Odonata is one of the most well known insect groups in South America. Within the order
Odonata there are two main suborders, encompassing dragonflies and damselflies. It is composed
of 36 species in 18 different genera, with 60% of the species and 40% of the genera endemic to
the region. We selected two damselfly species, Rhionaeshna variegata and Cyanallagma
interruptum to perform the first phylogeographic study of Patagonian Insects. They are the most
widespread damselfly species of the region, and are found in diverse habitats with varying
dispersal ability. Phylogeography is a strong tool in exploring the complex process of historical
diversification, however only moderated attention has been given to Patagonia from a
phylogeographic perspective. The species we selected are very suitable for this type of study due
to their abundance, the ease of collecting and their wide range of dispersal capability.
Methodology
Sampling and preliminary analysis:
From 2013 to 2015 we conducted extensive fieldwork in Patagonia (from northern Neuquén to
southern Santa Cruz) sampling in forest and grassland habitats. We fixed the dragonfly
specimens in 96% ethanol and stored them at -80°C in Brigham Young University (BYU, UT
USA) until DNA extraction. The vouchers are stored in the Insect Genomics Collection (IGC),
M.L. Bean Museum, at BYU. In order to conduct a preliminary phylogeographic analysis we
selected nine populations of Aeshnidae and Coenagrionidae. Based on those results we selected
Coenagrionidae to conduct further analysis with additional populations.
DNA sequencing
We extracted genomic DNA from muscle tissue, dissected from the leg, using the Qiagen
DNeasy protocol for animal tissue. The molecular data set is comprised of the mitochondrial
COXI/II, and recently developed EPIC (exon-primed intron-crossing) nuclear genes PRMT,
CDC5 and AgT (Ferreira et al., 2014). The primers and amplification protocols are summarized
in table 1. Sequences were generated using an ABI 3100 capillary sequencer at the BYU DNA
Sequencing Center. All sequences were edited manually and aligned using MUSCLE, as
implemented in Geneious® software.
Results
Our preliminary analysis of Coenagrionidae showed a clear genetic structure, whereas Aeshnidae
did not and was excluded from further analysis. The results are consistent with greater dispersal
ability among the larger aeshinds as compared to the smaller coenagrionids. Further analysis,
using COXI/II, PRMT, CDC5, and AgT, was performed
with the addition of eleven additional populations of C.
interruptum (20 populations and 54 specimens total) that
cover most of the species distributional range (Fig. 1) in
Patagonia.
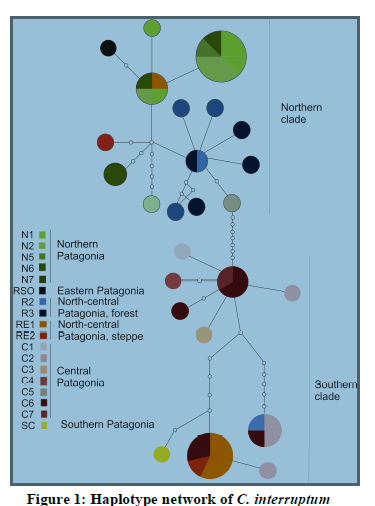
The mtDNA results for COXI/II show that C.
interruptum consists of two major clades, one distributed
in Northern Patagonia and one in Southern Patagonia,
with considerable genetic differences. Individuals from a
small region located in between these two regions in the
Río Negro grassland that have haplotypes recovered from
both clades.
Within the major Northern clade, there are two
subclades. The first includes haplotypes from Neuquén
and two additional haplotypes from the Río Negro
grassland. The second Northern clade includes
haplotypes solely from the Río Negro forest.
The major Southern clade also contains two subclades, one consisting of six haplotypes from
Chubut (C1-7 in Fig. 1), and another composed of four haplotypes, also represented in Chubut
(C), the Río Negro grassland (RE), Neuquén (N) and Santa Cruz (S). This last clade includes
populations from the largest distributional range, but does not contain populations from Northern
Patagonia (Neuquén). The nuclear genes showed a high level of heterozygosity but low levels of
genetic structure with absent geographic structure signals.
Discussion
The pattern revealed by the mitochondrial genes is concordant with a separation of the ancestral
population in the past and a posterior secondary contact in intermediate localities. It is probably
that events during the Pleistocene (glaciation cycles and associated changes in climate and/or
other geological events) interrupted gene flow among the northern and southern populations.
The incongruence between the mitochondrial and nuclear markers allows us to explore two nonexclusive
main hypotheses: 1) females may have lower dispersal capacity or high philopatry
compared to males, and 2) the temporal separation between the two populations was relatively
brief, providing enough time for mitochondrial markers to differentiate genetically, but not
sufficiently long enough to be reflected by the more constrained evolutionary rates of the nuclear
genes.
Conclusion
On average, female insects are heavier than males, and thus their flight capacity could be
reduced. However, as of yet there is no information available in the literature to sustain this
statement or a higher philopatry in females. On the contrary, there is evidence of Patagonian
vertebrates with genetic structure in the mitochondrial genes, but not in the nuclear genes. This is
most likely due to the different mutational rates of these genes.
