Klabacka, Randy
Phylogeny and species boundaries in the “flying dragons” of the Draco maculatus species
complex (family Agamidae)
Faculty Mentor: Dr. Jack W. Sites Jr., Biology Department
Introduction
Indo-Burma, comprising most of Mainland Southeast Asia east of India and north of Peninsular
Malaysia, is a biodiversity hotspot within Southeast Asia that has been called a contender for the
“hottest of hotspots” (Mittermeier, 1999). A remarkable majority of this species diversity
remains hidden from human recognition in two areas: (1) undiscovered in natural habitat, and (2)
conspicuously masquerading under known taxa. While solutions to the former require continued
field work, solutions for the latter require further study of already identified taxa for hidden
diversity therein. Indo-Burma is the endemic home to the “spotted flying dragon” (Draco
maculatus; Gray, 1845), and includes the entire range for this taxon. A diagnostic character of
the genus Draco (Family Agamidae) is their patagia– epithelial membranes supported by
elongated thoracic ribs that can be extended voluntarily to generate lift as they glide from one
tree to another. This region of the Old World tropics is renowned for discoveries of many new
species, yet no study has focused on resolving relationships and species boundaries in D.
maculatus. Cryptic species and hidden diversity can be revealed through molecular analyses
(Jörger, 2013), and this approach has doubled the known species richness in the genus Draco
since 1999 (Honda, 1986; McGuire, 2001). Due to the widespread range of D. maculatus and the
color and pattern differentiation among subspecies, we hypothesize that this alleged species is
actually a species “complex” currently masquerading under the nomen D. maculatus.
Methodology and Results
We obtained a total of 111 D. maculatus tissue samples from seven museums/collections for use
in this study. Many of these were field collected by members of our team, while others were
found on Genbank or VertNet and formally requested (see acknowledgments for institute
names). Although we were able to obtain sampling coverage for most of the D. maculatus range,
gaps remain for which no tissues are available. After extracting genomic DNA from liver
samples, we amplified and sequenced three mitochondrial genes (ND2, 12S, 16S) using a double
stranded polymerase chain reaction (as described in Davis et al, 2016). After concatenating our
mitochondrial sequences (consisting of data from all individuals), we Used IQ-TREE (Nguyen,
2015) to run an ultrafast bootstrap partition-by-gene Maximum Likelihood analysis with 1000
replicates. This generated a mitochondrial gene tree from which we subsampled 85 individuals to
include in an expanded dataset with three nuclear loci (BDNF, CMOS, and PNN). This produced
a matrix of over 4,000 nucleotide base pairs, and with the methodology described above
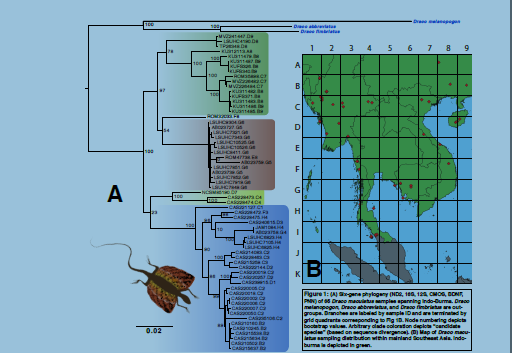
(substituting a full-bootstrap method for ultrafast), generated a multi-gene tree (Fig. 1A).
Discussion & Conclusions
Our phylogeny recovers four well-resolved and strongly divergent lineages within D. maculatus.
We therefore recognize this group as a species complex and hypothesize that four distinct species
are present within the nominal taxa D. maculatus. Whether this divergence is significant enough
to identify undescribed species will depend on rigorous examination of morphological
characters, so we refer to these clades as “candidate species” requiring further study. Further, we
noted the significant sampling gap within the geographic distribution of this taxon (Fig 1B).
Thailand includes three of the four type localities of the recognized D. maculatus subspecies:
Acknowledgements
We thank the California Academy of Sciences (CAS), Kansas University (KU), the La Sierra University
Herpetological Collection (LSUHC), the Museum of Vertebrate Zoology (MVZ), the National Museum of Natural History (USNM), the North Carolina Museum of Natural Sciences (NCSM), the Royal Ontario Museum (ROM), and other collaborative contributors (Dr. Lee Grismer, Dr. Jimmy McGuire).
References
1. Davis, Hayden R., L. Lee Grismer, Randy L. Klabacka, Mohd Abdul Muin, Evan SH Quah, Shahrul Anuar, P. E. R. R. Y. L JR, and Jack W. Sites. “The phylogenetic relationships of a new Stream Toad of the genus Ansonia Stoliczka, 1870 (Anura: Bufonidae) from a montane region in Peninsular Malaysia.” Zootaxa 4103, no. 2 (2016): 137-153.
2. Gray, John Edward. Catalogue of the Specimens of Lizards in the Collection of the British Museum. order of the Trustees, 1845.
3. Honda, Masanao, Hidetoshi Ota, Mari Kobayashi, Jarujin Nabhitabhata, Hoi-Sen Yong, and Tsutomu Hikida. “Phylogenetic relationships of the flying lizards, genus Draco (Reptilia, Agamidae).” Zoological science 16, no. 3 (1999): 535-549.
4. Jörger, Katharina M., and Michael Schrödl. “How to describe a cryptic species? Practical challenges of molecular taxonomy.” Frontiers in Zoology 10, no. 1 (2013): 1.
5. McGuire, Jimmy A., and Kiew Bong Heang. “Phylogenetic systematics of Southeast Asian flying lizards (Iguania: Agamidae: Draco) as inferred from mitochondrial DNA sequence data.” Biological Journal of the Linnean Society 72, no. 2 (2001): 203-229.
6. Mittermeier, Russell A., Norman Myers, Cristina Goettsch Mittermeier, and P. Robles Gil. Hotspots: Earth’s biologically richest and most endangered terrestrial ecoregions. CEMEX, SA, Agrupación Sierra Madre, SC, 1999.
7. Nguyen, L.T., Schmidt H.A., von Haeseler A. & Minh B.Q. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol, 32:268–274. doi:10.1093/molbev/msu300