Robert Swenson and Dr. David Belnap, Chemistry and Biochemistry
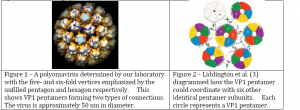
Polyomaviruses (PVs) are viruses that can cause tumors in mammals and fatal diseases in birds. Knowles et al. (2) estimated that over 90% of the human population is infected with a polyomavirus, but these infections are usually dormant, and only show symptoms in patients with compromised immune systems. PVs normally form a capsid with icosahedral symmetry whose structure is known (Figure 1). In this structure, the major capsid protein (VP1) forms a stable pentamer with the C-terminal arms of the proteins extending outwards. VP1 pentamers (filled pentagons, Figure 1) interact with five other pentamers on some vertices, and with six neighboring pentamers on other vertices. The C-terminal extensions coordinate to hold neighboring pentamers together in the overall capsid structure.
VP1 has shown an unusual flexibility and is able to form several different structures from the same stable pentamer substructure including small and large icosahedral shells as well as helical tubes and octahedral particles. This variation would be like assembling the same pieces of a jigsaw puzzle in four different ways, to see four different pictures. To form an octahedral (cubic) structure, VP1 must change its bonding to coordinate four neighboring pentamers (edge to edge). We proposed to determine what specific changes VP1 undergoes in order to form octahedral shells with the unique four-fold relationship.
Robert Garcea, University of Colorado, provided us with a plasmid containing the gene for the main viral capsid protein (VP1). He also provided us with a protocol for growing large quantities of the protein in E. coli cells. Following this protocol we were able to grow and purify sufficient quantities of VP1 to form octahedral particles.
The procedure for inducing VP1 pentamers to form octahedral particles was outlined by Salunke et al. (4). Following their procedure I dialyzed my purified sample of VP1 against three buffers:
1- pH 7.2, with EDTA, 2-mercaptoethanol, glycerol
2- pH 8.5, with EDTA, 2-mercaptoethanol, glycerol
3- pH 7.2, with CaCl2 , glycerol
The EDTA, and 2-mercaptoethanol were used to prevent premature aggregation of VP1. The CaCl2 in the third buffer is necessary for particle formation and is incorporated into the capsid. We used negative stain electron microscopy to visualize the formation of these particles. In negative staining specimens are stained using a heavy metal so that they can be visualized in the electron microscope. After successful formation of octahedral particles, we imaged them using cryo-em.
Cryo-em is a technique for visualizing biological specimens in a native or near-native state. To prepare the sample for cryo-em we placed a small amount of the octahedral particles on a holey-carbon cryo-EM grid, and froze the sample very quickly by submerging the grid in liquid ethane. The grid is then placed in the electron microscope and images are taken on photographic plates.
For most of the year we were unsuccessful in imaging the octahedral particles by cryo-em. Preparing the holey-carbon grid requires some practice because the grids are very fragile and frost often forms on the grids during transfer preventing us from being able to image them. Our time on the electron microscope has also been limited because the microscope has been down several times during the year, as well as our sample holder for the microscope has been out for repair several times. Last week we received our sample holder back and found that it was functioning correctly. We were able to prepare a sample with good ice. We imaged the sample and have developed the images.
We are now in the process of digitizing the images using a commercial grade scanner. As the images are scanned we will assess image quality using Fourier transform methods (1) and only suitable images will be used further. Images of octahedral particles will be extracted using freely available software. The 3D “reconstruction” will be generated using an iterative method that aligns and averages the images.
Once the model is fully refined I will use the previously determined pentamer coordinates from the crystal structure of icosahedral murine polyomavirus (5) to model how interactions in the octahedral particles differ from those in the icosahedral particles. We hypothesize that the majority of the changes in the structure of VP1 will be in the C-terminal arm and that the core of the capsomere will be largely conserved.
Sources
- Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-Angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745-7755.
- Knowles, W. A. 2006. Discovery and epidemiology of the human polyomaviruses BK virus (BKV) and JC virus (JCV). Adv. Exp. Med. Biol. 577: 19-45
- Liddington, R. C., Yan, Y., Moulai, J., Sahli, R., Benjamin, T. L., and Harrison, S. C. 1991 Structure of simian virus 40 at 3.8-Å resolution. Nature 354: 278-284.
- Salunke, D. M., D. L. D. Caspar, and R. L. Garcea. 1989. Polymorphism in the Assembly of Polyomavirus Capsid Protein VP1. Biophys. J. 56:887-900.
- Stehle, T., Harrison, S.C. 1997. High-resolution structure of a polyomavirus VP1-Oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139-5148.
