Michael Lydiksen and Dr. Adam Woolley, Chemistry and Biochemistry
Introduction
Imagine having the ability to control the placement of atoms with reasonable speed and accuracy. Having this capacity naturally brings with it incredible technological advancements in areas such as integrated circuits and biochemistry. An exciting process known as dip-pen nanolithography (DPN) that I have been actively perfecting over the past year gives researchers the ability to control the placement of individual atoms at a desired location on a substrate. I have presented some of my results last spring in BYU’s annual College of Physical and Mathematical Science Spring Research Conference.
Problem
As part of an interdisciplinary team of physicists, chemists, and engineers at BYU known as the ASCENT group we are trying create nano-circuits out of DNA. To do this we take a long scaffold strand of single stranded DNA and with the appropriate staple strands we can actually fold DNA into desired shapes known as DNA origami. We eventually want to create a circuit shape and connect multiple circuits in series. However, one of the main problems with this process is that the DNA origami aligns itself in random patterns. (Figure 1) DPN is one of the ways that has been proposed to orient the DNA at desired locations on a surface
Methods
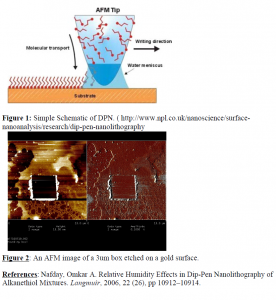
An atomic force microscope (AFM) is used to perform DPN. An AFM is a precision instrument capable of imaging at the nanoscale. A small mechanical probe a little over 3mm long called a cantilever is placed in the AFM and is tuned to oscillate at its resonant frequency. The laser is aligned on the tip of the cantilever and the cantilever begins probing the surface. We can think of the cantilever as a pen that can be dipped in ink and then used to write on paper just like a quill. In my experiments the ink is octadecanethiol and the substrate is an atomically flat gold surface. Thiols form covalent bonds with gold and arrange themselves into rigid structures known as self-assembling monolayers (SAM). To perform DPN, the cantilever is dipped into a really tiny drop of thiol solution and allowed to sit there for 20 seconds. Once the tip has been thoroughly coated in a thiol solution then it is placed in the AFM and a humidity chamber is placed over the AFM. Humidity is one of the key factors affecting DPN. When the tip is close to the surface a meniscus is formed and the size of this meniscus is linearly related to humidity. The optimum relative humidity is between 30-40% (Saha) but this can greatly vary due to other factors such as tip speed and substrate conditions.
Results
My initial approach to the problem at hand was to become proficient in the use and operation of the AFM. Once I became proficient we bought atomically flat gold surfaces and I began some initial DPN experiments. One of the first problems that I encountered was that instead of depositing thiol on my gold surface I was bringing the tip too close to my surface and I was actually etching my surface. (Figure 2) To adapt to this problem I increased the deflection setpoint on the AFM which essentially raises the tip slowly from the surface. However, this process seemed to be very selective and sometimes I would see a box on one setpoint value and then the next time with the same setpoint value I wouldn’t see it. After a long time I decided to switch to a much gentler imaging mode known as tapping mode. Instead of painting boxes on my surface I decided to just try simple diffusion of thiol onto my gold surface. I am currently working on perfecting this process and while I have seen what appear to be diffusion dots at times, further work is needed to streamline my process so that DPN can be done quickly and efficiently.
Discussion
I have set out to perfect a technique known as dip-pen nanolithography that is one of the available options that would allow us to systematically place DNA origami at desired locations on a surface. While I have made progress in this endeavor, further research is needed to finish my project. Once I can show that I can place thiols at desired locations on my surface without etching then my future experiment will be to repeat the same process with thiolated DNA that is complementary to my DNA origami. The DNA origami will bind with the thiolated DNA and the excess DNA that is not bound to the surface will be washed away.