Laura Oh and Josh Andersen, Department of Chemistry and Biochemistry
Introduction
SOD1 is overexpressed in many cancers such as lung adenocarcinoma, non- small-cell lung cancer, and 70% of primary breast cancers. Traditionally, SOD1 is known as an antioxidant in the human body, but only 1% of SOD1 expressed in cells is necessary to keep superoxide radicals below cytotoxic levels, suggesting that SOD1 may have undiscovered functions. One of the known trademarks of cancer is the Warburg Effect defined by up-regulated glycolysis and down- regulated respiration. It is thought that this unique mode of metabolism promotes cancer cell growth. A recent study demonstrated that SOD1 functions to repress respiration through the casein kinase pathway, thereby possibly linking SOD1 to the Warburg Effect and explaining why SOD1 is overexpressed in cancer. We recently found that the 123rd amino acid of SOD1 (a lysine, K123) is modified by acyl groups. K123 is found in the electrostatic loop of SOD1, which is a critical domain responsible for binding copper, a cofactor of SOD1 that allows it to be active. These studies revealed that when SOD1 is acyl modified at lysine 123, it loses its ability to suppress respiration, which would lead to a loss of the Warburg Effect and decreased proliferation of cancer cells (Figure 1). While the effect of acyl modification on SOD1 is clear, we do not yet know how acyl modification disrupts this function of SOD1
Methodology
- Identify proteins that bind to wild type SOD1 but not the acyl-modified SOD1.We used co-immunoprecipitation techniques and proteomics tools to look for proteins that interact with wild type SOD1 to mediate its effect on respiration.
- Determine whether acetylation affects binding of copper to SOD1. We used co-immunoprecipitation and Western blotting to determine if the acyl-modified mimics of SOD1 can bind copper and dimerize properly.
Results
Proteomics on SOD1
We were able to perform pull down experiments with SOD1 in preparation for proteomics. We found a few interesting interactors, a few of which have been shown previously including CCS and p62, and also a few novel interactors such as TCP4, PARP1,IF1AX, HMGB1 and HMGB2.
Determination on acetylation affects in binding of copper to SOD1
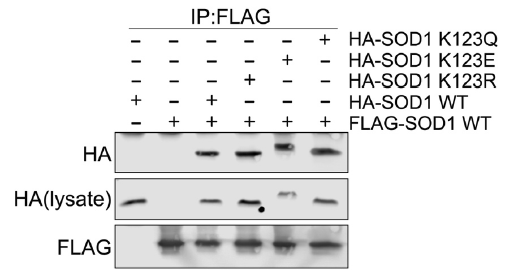
We analyzed the affinity of copper binding to the acyl-modified mimics of SOD1 by looking at dimerization of the acyl-modified mimics with wild type SOD1. Figure 1 shows that acyl modified SOD1 dimerizes equally well with wild type SOD1 as wild type SOD1 dimerizes with itself, which means that the acyl-mimics of SOD1 do not lose their affinity to bind copper since binding copper is necessary for dimerization. This implies that SOD1’s inability to suppress respiration when it is acylated is not due to a decreased affinity to bind copper or dimerize.
Figure 1 – We overexpressed WT Flag-SOD1 along with WT and acyl-mimics of HA-SOD1. We immunoprecipitated WT Flag-SOD1 and looked for a change in the interaction of WT HA-SOD1 vs the acyl-mimics of HA-SOD1.
Discussion
Our hypothesis was that acyl modification inhibits SOD1’s activity by disrupting SOD1 binding to Cu and/or disrupting SOD1 interactions that are critical for suppressing respiration. We found out that SOD1 loses its ability when it is acyl- modified. For a future aim we want to look at whether acyl-modification causes SOD1 to lose interaction with any of the interactor we found through proteomics. We will use western blot to validate the interaction between wild type SOD1 and mutant SOD1. Through western blot, it will tell us whether it will disrupt binding copper to SOD1 or not.
Conclusion
Although we are still in the process of understanding the mechanism of SOD1’s function in cancer proliferation, we were successful in laying out the background on how SOD1’s inability to suppress respiration by acyl-modification is not due to a decreased affinity to bind copper or dimerize. We are looking forward to perform more experiments with interactors we found through proteomics.