Andrew Dearden and Adam Woolley, Chemistry & Biochemistry
With countless technologies that rely on microprocessors, there is a great need for increasingly smaller microelectronic components. Traditionally, manufacturers have employed a “top-down” approach to build microelectronics – that is, they have attempted to scale functional microcircuitry components down in size, hoping for similar performance in a smaller circuit. This top-down approach is expensive, often costing manufacturers hundreds of dollars per part. A much more cost-effective approach to microelectronics has emerged over the past decade in the field of molecular science: that is, to develop circuitry from the “bottom-up” using macromolecules such as deoxyribonucleic acid (DNA). Scientists have now devised methods to fabricate conductive nanowires using DNA templates. Because DNA chemistry is well-understood, easily controlled, and inexpensive, it has great potential to become the new standard for nanowire fabrication.
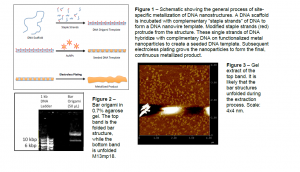
In my research, I construct DNA-templated nanowires by incubating the single-stranded viral DNA, M13mp18 (also known as the “scaffold” strand) with several hundred smaller strands of DNA also known as “staple strands.” These artificial DNA strands are designed to complement with the scaffold strand such that when the staple strands attach, they fold the scaffold strand into a desired “bar” shape. Successful DNA folding is verified under an atomic force microscope. In addition to folding the scaffold strand, the staple strands are also designed to serve as attachment sites for gold nanoparticles (AuNPs). AuNPs are made to attach at the specific staple strand sites by activating AuNPs with thiolated DNA strands that are complementary to the DNA at the attachment points. Thus, simple incubation of activated AuNPs and the DNA nanostructure will yield a nanoparticle-DNA complex dotted (or “seeded”) with AuNPs. This seeded structure is treated with electroless plating to form the final nanowire. The entire fabrication process is shown in Figure 1.
One of the core issues in DNA nanowire fabrication arises during electroless plating: any excess AuNPs not attached to the seeded DNA structure will be plated too, thereby contaminating the surface with extra nanoparticles and lowering the yield of AuNP deposition on the seeded DNA structure. The result of this problem is a non-uniform nanowire that cannot reliably transport charge. Theoretically, removal of contaminant AuNPs prior to electroless plating will improve metallization on the nanowires. For my ORCA project, I investigated how gel electrophoresis might be used to remove excess AuNPs from solution prior to electroless plating, as gel electrophoresis is a well-established method for high quality purification of biomolecule mixtures. Specifically, I employed a proof-of-concept approach by showing that DNA nanostructures can be successfully extracted from an agarose gel following gel electrophoresis.
After successful synthesis of the DNA bar structure, 50 μL volumes of 5 nM bar templates were mixed with 2 μL loading dye and 5 μL. The mixtures were loaded into 0.7% agarose gels. Ethidium bromide was used both in the agarose gel and in 0.5x TBE running buffer as a staining agent to visualize the DNA on the gel. Each gel was run for approximately two hours at 70 V. The samples were extracted according to the protocol accompanying the Qiagen QIAquick Gel Extraction Kit. A 1 kb DNA Ladder from GeneRuler™ was used for control purposes.
Each gel invariably resulted in the formation of two distinct bands as shown in Figure 2. Subsequent control gels and AFM analyses confirmed that the top band is the folded bar nanostructure, whereas the bottom band is unsuccessfully folded scaffold DNA.
I performed Qiagen’s extraction protocol on numerous gel samples. I made a pair of adjustments that seemed to substantially increase the yield of extracted DNA. First, I found that incubating the extracted DNA with EB buffer for several hours (an average of four) before elution increased recovery. Heating the EB buffer to 50 °C before incubation also increased DNA recovery.
Figure 3 shows an AFM image of the top band extract. Several DNA structures can be identified. It is curious that none of these structures resemble the DNA bar structure when compared to control images. I hypothesize that the DNA looks globular and unstructured because the bar nanostructures are subjected to high temperatures during the extraction process. High temperature and other stresses likely compromised the structural integrity of the bar structure, so the DNA structures in Figure 3 are partially unfolded.
Further tinkering with extraction methods will be required in order to efficiently and consistently recover the bar nanostructures from agarose gels. After this process is refined and we know that we can recover uncompromised bar structures in high yield, we can actually seed the bar structures with AuNPs and determine whether removal of excess AuNPs prior to electroless plating does indeed increase metallization yields on the DNAtemplated nanowire.