Shankar Parajuli and Dr. Allen Buskirk, Chemistry and Biochemistry
Ribosomes are molecular machines that synthesize all the proteins for all living cells. Ribosomes sometimes stall and cannot complete the synthesis of a given protein. Some peptides interact with the ribosome during their own translation and induce stalling.1 The SecM and TnaC peptides in E. coli, for example, have been found to play a key regulatory role in translation of genes found on the same mRNA.2
To date, stalling peptides have been identified one at a time through efforts to understand the mechanism of individual genetic switches. The goal of our research was to identify many new stalling peptides and through additional characterization, to further understand how they interact with the ribosome to induce stalling. Here, we report the validation of this selection and the identification of several novel stalling peptides.
A new bacterial two hybrid system was designed as a genetic screen to identify stalling peptides. This selection linked stalling to cell survival by tying stalling to the transcription of an essential gene required for histidine biosynthesis. In this system, when the ribosome stalls at the the C-terminus of the DNA-binding protein cI, a natural bacterial molecule called transfer messenger RNA (tmRNA) rescues the stalled ribosome. This rescue leads to the addition of a modified tmRNA tag (AANDENYALDD) at the end of the cI protein. After the tag gets added, the first seven residues of the tag (AANDENY) then interact with its protein partner SspB fused to the RNA polymerase alpha (RNAP).3 The SspB-RNAP fusion used in our selection does not contain the ClpXP binding region and hence does not degrade the protein. The interaction of the tmRNA tag with SspB recruits the transcriptional machinery to actively transcribe the HIS3 gene off of a weak promoter (Pweak). If no stalling occurs, transcription of the gene is not enough for cellular survival. Hence, bacterial cells undergoing ribosome stalling followed by tmRNA rescue will make histidine and survive on media that lacks histidine. A brief summary of this approach is shown in Figure 1.
After validating the selection, pBT-library was designed by inserting 20 random codons (no known stalling sequence) at the end of cI protein. The pBT-library plasmid was then incorporated into competent cells (containing two plasmids to express required proteins), grown overnight, and plated on both-rich media and selection media plates. About 160 of the surviving colonies were picked from the selection plates, and the pBT plasmid DNAs were sequenced. Upon analysis, we found 3 classes of sequences. Class I included sequences that survive selection due to the presence of SspB binding motif (similar to AANDENY) in the random sequence of library. These peptides probably do not induce stalling and were not further characterized. Class II included sequences with Pro-stop. As evident in the case of TnaC and EP-stop, peptides ending in Pro-stop are well characterized members of stalling peptides. 4, 5Class III included all other sequences from cells that survive selection for reasons unknown and were hence of interest to us.
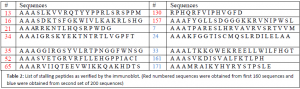
We then used class III peptides to look directly for addition of the tmRNA tag to the lamba cI portein to verify that the survival was a result of tmRNA rescue and tagging. Immunoblot was performed by the use of the primary antibody specific to DD-tag (end of tmRNA tag). Of the samples used in this immunoblot, peptides numbered 13, 16, 21, 34, 35, 52, 65,130, and 157 showed cI tagging (labelled in red in Table 1).
In an attempt to minimize false positives and maximize output from first round of screening, second generation of selection was performed by including streptomycin, in addition to histidine, into the assay. In this second round of screening, we first plated the cells on selective media plates and then transferred the surviving colonies into another set of selection plates containing streptomycin and allowed cells to grow. We picked 200 colonies from these new selection plates and sequenced the pBT plasmid DNA. Results similar to that obtained in first round of screening were observed. Further subjection of interesting peptides to immunoblot, by the use of DD-tag antibody, eliminated several false positives leaving us with only 5 stalling peptides (labeled in blue in Table 1).
From the results obtained, we found that most of the stalling peptides were sequences ending with Pro-stop. These are well characterized classes of stalling sequences. In addition, the majority of peptides that survive our selection do not show tmRNA tagging, as we discovered after immunoblot assays that were performed using the DD-tag antibody. This led us to believe that there were several other reasons for cellular survival other than tmRNA tagging that our selection assay failed to address. An attempt of minimizing false positives by the use of streptomycin resistance in second screening did not yield better results. We believe that a better way of maximizing the output would be to design a random sequence library in such a way that proline codons are avoided before the stop codon. Nonetheless, we found 14 new sequences that induce tagging by tmRNA- 9 from first and 5 from second round of screening that are interesting and worth characterizing. We conclude that stalling peptides are many and do not show significant sequence similarity. Their thorough understanding will render us information as to why stalling is important in vivo.
Of many challenges faced during the course of the project, reproducibility was the greatest. This problem directly tied with the preparation of selective media. We found that cells were very sensitive to the amount of ingredients used and the temperature they were grown at. So, the optimum amount of ingredients was determined by running several rounds of titrations and plating experiments to find the best possible environment for cells to grow.
The future of this project is to characterize all 14 newly found stalling sequences. The characterization process involves 3 steps: toeprinting assay to detect the exact spot of stalling, mutagenesis to determine the essential residues in the sequences of 20 amino acids, and mass spectrometry to determine the exact spot of tagging by tmRNA.1,6 These characterization experiments are currently being done by a graduate student Chris Woolstenhulme and 2 other fellow undergraduates- Diana Valverde and Levi Lowry. The outcome of these experiments will help us understand the biological significance of ribosome stalling in newly found peptides.
References
- Tanner, D. R.; Cariello, D. A.; Woolstenhulme, C. J.; Broadbent, M. A.; Buskirk, A. R. Genetic Identification of Nascent Peptides That Induce Ribosome Stalling. J. Biol. Chem. 2009, 284, 34809-34818.
- Gong, F.; Yanofsky, C. Instruction of translating ribosome by nascent peptide. Science 2002, 297, 1864-1867.
- Dougan DA, W.-B. E., Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell. 2003, 12, 373-380.
- Christopher S. Hayes, B. B. a. R. T. S. Proline Residues at the C Terminus of Nascent Chains Induce SsrA Tagging during Translation Termination. The Journal of Biological Chemistry 2002, 277, 33825-33832.
- Ito K, C. S., Poqliano K Divergent stalling sequences sense and control cellular physiology. Biochem Biophys Res Commun 2010, 1, 1-5.
- Gould PS, B. H., Easton AJ Translation toeprinting assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques 2005, 38, 397-400.