Matthew McDowell and Dr. Allen Buskirk, Department of Chemistry and Biochemistry
Abstract
Ribosomes are large complexes made up of ribosomal RNA and proteins and are the site of protein synthesis. However, ribosomes occasionally run into problems during this process and will stall during translation. Fortunately, two molecules known as tmRNA and SmpB are made by cells in order to free stalled ribosomes. We sytematically mutated G517, C518, C519 and A520 in the ribosome to see if binding fidelity decreases, and to see if these bases affect the activity of tmRNA/SmpB in ribosomes during ribosome rescue. This assay however, gave us no informative results because we were only looking at endpoints of its rescue.
Introduction
Ribosomes are large complexes made up of ribosomal RNA and proteins and are the site of protein synthesis. Ribosomes are made up of two subunits that bind an mRNA template with the help of initiation factors to form the ribosome complex. Ribosomes work by reading the codons on the mRNA and matching them up with other molecules called tRNA. A tRNA is a molecule that can base pair with a sequence located on the mRNA template called a codon. This is a process known as translation and is used in cells in order to produce proteins necessary for life. Normal translation in prokaryotic cells occurs with the aid of two ribosomal subunits, the 30S and 50S subunits. The 30S subunit contains a decoding site where the codon-anticodon interaction is read. The 50S subunit is responsible for peptidyl transfer and is the location of hydrolysis reactions. Ribosomes contain three tRNA binding sites. The A site is the amino-acyl, the peptidyl transfer site is the B site, and the exit is the E site. Nascent peptides are formed as a charged tRNA enters the A site and base pairs with mRNA with the help of elongation factor EF-TU . Then with the help of peptidyl-tRNA in the P site and elongates the peptide. This reaction is followed by translocation which is catalyzed by elongation factor EF-G and initiates the movement of the peptidyl-tRNA and the deacylated tRNA into the P and E sites1. (Details of translation can be reviewed from Sauer et. al.)2. However, ribosomes occasionally run into problems during this process and will stall during translation.
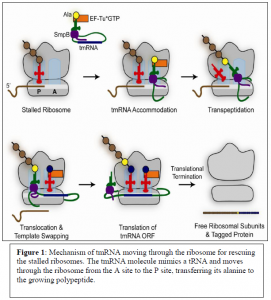
Fortunately, two molecules known as tmRNA and SmpB are made by cells in order to free stalled ribosomes. tmRNA and SmpB recognize ribosomes containing an empty A site which serves as a substrate for the tmRNA/SmpB to come in and rescue stalled ribosomes by sequestering the two subunits. Figure 1 shows the mechanism in which tmRNA moves through stalled ribosomes in order to separate the two subunits from the mRNA template. The tmRNA molecule acts as though it were a normal tRNA and moves through the ribosomes from the A site to the P site and transfers an alanine to the growing poly peptide. The tmRNA has a tail portion which then acts as an mRNA template. This template codes for a protein tag that will be attached to the nascent peptide. The added peptide tag signals for protein degradation and allows recycling of the ribosome subunits.
Adapting Dr. Allen Buskirk’s conceptual model, we have come up with a hypothesis and a conceptual model of how and where tmRNA binds to the ribosome. We have hypothesized that once tmRNA binds to SmpB, the SmpB protein then binds to the ribosome on the 520 loop of the 16S rRNA in the A site of the ribosome. Testing the validity of this conceptual model will help us to answer the question of how the tmRNA/SmpB rescue complex is recognized by the stalled ribosome.
Results and Conclusions
In order to test this hypothesis we systematically mutated G517, C518, C519 and A520 to see if these bases affect the activity of SmpB in ribosomes during ribosome rescue. First, we wanted to determine the activity of our mutant ribosomes during normal translation. In order to do this, we tested for the activity of the mutated ribosomes through experiments using the reporter gene glutathione S-transferase (GST). The reporter Gene GST inserted was modified to stall in cells and tmRNA was also modified to add a his-tag to stalled proteins as opposed to the normal degradation tag. When this modified GST was expressed, the modified tmRNA rescued the ribosome stalled on GST messages. We could then measure the level of rescue of GST by tmRNA through western blot. However, the western blots did not show any differences in tagging for those ribosomes that were still functional.
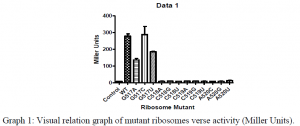
Additionally, we conducted Beta-Galactosidase Assays in which we measured the the amount of ONPG (ortho-Nitrophenyl-β-galactoside) that was hydrolyzed by the LacZ gene product when induced. The results from this assay are contained within Graph 1. Unfortunately, the activity of our mutated orthogonal ribosomes, especially the mutated 518, 519, and 520 bases appeared to have denatured insomuch that the ribosomes lacked any appreciable activity. Even though the ribosome mutants appeared to be dead, we were going to use the ability to purify mutant ribosomes to try to work around this problem, but Ramakrishnan recently published a paper which solved the structure of tmRNA/SmpB in the ribosome. His structure showed that there was no apparent interaction between the 520 loop and the tmRNA/SmpB complex. The results from Ramakrishnan’s paper3, and the lack of activity of the mutated ribosomes shown from our Miller Assays has lead us to investigate new methods and hypothesis to answer our question of how the tmRNA/SmpB rescue complex is recognized by stalled ribosomes.
Discussion
Investigation of the ribosomes of prokaryotic cells did not provide evidence of tmRNA/SmpB complex interaction with the 520 loop. The results obtained from the Miller Assays have lead us to believe that our ribosome complexes were not forming; suggesting the 520 loop is crucial for the ribosome in maintaining its correct conformation and function. The mutations on the 520 loop interfere with ribosome complex formation and cannot be reformed. We did not see any decrease in tagging in any of the mutants that expressed and the data from the Western Blots of the endpoints can be misleading. Additionally, because the results of Ramakrishnan’s data show no interaction between the 520 loop and the tmRNA/SmpB complex, there must still be interaction somewhere just not in the 520 loop. Because of these results, our lab under the direction of Dr. Allen Buskirk will now move on and test SmpB mutants that do not support rescue. We will then test in vitro if the mutated SmpB will still bind to the ribosomes, and at what point these mutants affect the decoding mechanism during ribosome rescue.
Methods
Ribosomes contain a Shine-Delgarno sequence which recognizes and binds to mRNA strands that are to be translated into a protein product. In order to run our Miller Assays, we used ribosomes that have a mutated Shine-Delgarno sequence; a method used and described by Kurt Fredrick4. Our orthogonal ribosomes would then only attach to specially designed mRNA strands that would base pair with the orthogonal ribosomes only. Our specially designed mRNA strands contained the LacZ gene then would be translated into a protein when induced. We then could test how much of the LacZ gene product was made by the orthogonal ribosomes by observing how much ONPG (ortho-Nitrophenyl-β-galactoside) was cleaved and turned yellow in color. The amount of cleaved ONPG was then measured using a spectrophotometer. In the western blot we used antibodies against his-tag and also against GST in order to normalize our data.
References
- Zaher, H.S and Green, R. Fidelity at the Molecular Level: Lessons from Protein Synthesis. Cell 136, 746-762, February 20, 2009.
- Sauer, R.T., and Moore, S.D.. The tmRNA System for Translational Surveillance and Ribosome Rescue. Annu. Rev. Biochem. 2007. 76:2.1-2.24
- Ramakrishnan, V, Neubauer, C, Gillet, R, and Kelley A.C. Decoding in the absence of a codon by tmRNA and SmpB in the ribosome. Science. 2012 Mar 16;335(6074):1366-9.
- Fredrick, K., McClory, S.P., Leisring, J.M., Qin, D.M.. Missense suppressor mutations in 16S rRNA reveal the importance of helicies h8 and h14 in aminoacyl-tRNA selection. RNA (2010), 16;1925-1934. Published by Cold Spring Harbor Laboratory Press. Copyright 2010 RNA Society.