Alicia Brighton and Dr. Julianne Grose, Department of Microbiology and Molecular Biology
Introduction
Fireblight is a disease, caused by the bacteria Erwinia amylovora, that can infect some fruit trees, including apple and pear trees. When an infection occurs, the effects can be devastating—any infected part of the tree must be removed and destroyed, sometimes resulting in loss of the entire tree. Furthermore, in the warm and moist conditions typical of springtime, the spread of fireblight can be nearly impossible to control with current management methods.1
My project was aimed at the development of an effective, as well as natural, organic, and nontoxic treatment for fireblight. This treatment involves harnessing the natural enemies of bacteria: viruses called bacteriophages, or phages. Applying to trees a mixture of phages that can kill E. amylovora could cure and prevent fireblight infections. But including different types of phages in the treatment is critical. Otherwise, a few bacteria that are resistant to the chosen phage could survive and reproduce, creating an infection completely resistant to that phage treatment. Using a cocktail of different phages that kill the bacteria in different ways creates a more reliable treatment.2
Objective
My original goal for this project was to take the DNA sequence for one E. amylovora phage and use computer programs to predict all of its genes. I would then check the validity of my gene predictions by comparing the sizes of proteins expected (based on the predicted gene sizes) to the actual phage protein sizes as determined by a method called mass spectrometry. The resulting phage genome, or map of the genes, would be useful because it could be studied and compared with other phage genomes to determine which other phages the phage could be combined with to create an effective fireblight treatment.
Unfortunately, I had problems and delays with the preliminary step: getting a DNA sequence. Our lab has been sequencing phage DNA routinely for three years, but these E. amylovora phages came with unexpected challenges. In the original sequencing attempt, none of the E. amylovora phage DNA samples produced sequences. I had to start almost back from the beginning to try and produce better DNA samples. I ended up doing a lot of troubleshooting to produce enough phages to get enough DNA. Because of the setbacks, my goal narrowed and became simply to obtain DNA samples of sufficient quality and quantity for sequencing.
Process
Phage Culture
The first step in getting a lot of phage DNA for sequencing is to get a lot of phage. I started with lower-concentration solutions of phage and used two methods to amplify, or grow, the phage. The first of these methods was to grow some phage-infected bacteria on a plate of nutrient-rich agar. Phages kill bacteria by taking over their “machinery” and using it to produce more phages. So as some of the bacteria died, they lysed (broke open), releasing phage and producing small clearings called plaques on the plates that were otherwise covered with growing bacteria. I could then harvest the phage from the plate. However, I found that I was not able to get enough phages using this method. The second method I tried was liquid culture. For this method, instead of growing the phage-infected bacteria on a plate, I grew them in a flask of liquid nutrient broth. After two to three days, a viscous, mucous-like build-up in the liquid culture indicated that bacteria cells were being lysed and a high concentration of phages had been produced. I could then filter the liquid to remove bacteria, leaving only the phage in solution. At first, I had a lot of problems with these liquid cultures getting contaminated. When that happened, the phages seemed not to infect and amplify properly. I had to remake broth, replace instruments, and modify my procedure until I eliminated the contaminants. I was finally able to produce a high concentration of each of the two phages I was working with.
DNA Extraction
With the high-concentration phage solutions, I was ready to extract the DNA. I first added a mixture of enzymes called nucleases to the phage solution to destroy any foreign DNA in the solution. Then I added a phage precipitate solution and centrifuged (spun) the solution to get all the phages into one pellet. After removing the liquid, I added a resin to the pellet to break open the phage and bind the phage DNA. I then used isopropyl alcohol to wash away the phage proteins, leaving only the resin bound to the phage DNA. I added TE buffer to remove the DNA from the resin and bring the DNA into solution.
Sample analysis
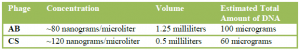
I determined the suitability of my DNA samples for sequencing by analyzing them in two ways. First, I measured the concentration and purity of the DNA samples using a machine (a spectrophotometer called the NanoDrop) that can measure the absorbance of different light wavelengths to determine the concentrations of various components in a single drop of a solution. Second, I compared my DNA samples to DNA samples of known purity and concentration using gel electrophoresis, a method where DNA samples are forced through a gel slab using an electric current and then the resulting bands of DNA are compared to one another.
Results and Conclusion
The above table shows the estimated amounts of phage DNA I extracted, based on the NanoDrop concentration measurements. The picture at right is the gel I ran, and the darker bands of the phage DNA samples show that their concentration is not as high as the known sample concentration. However, the amount and quality of each DNA sample is sufficient to be sequenced. By producing these DNA samples, I accomplished an important preliminary step in the characterization of these phages for the purpose of developing a phage treatment for fireblight. Currently, other students are using my sequence results to annotate the genome and my phage is being mass produced in liquid culture to spray on trees this October in our first trial of phage treatment for Erwinia. In addition, phage hunter students this year are using the protocols I developed. They have already isolated 15 new phages and will soon purify the DNA and sequence more.3