Wei Meng and Kim O’Neill, Department of Microbiology and Molecular Biology
Introduction
Cancer remains the second most common cause of death in the US, accounting for nearly 1 of every 4 deaths. Studies have shown that the early detection of cancer leads to better patient prognosis and a greater five-year survival rate. [1] Diagnostic and prognostic markers play a key role in classifying tumors and determining the best treatment plan for a patient. Among these biomarkers, Thymidine kinase 1 (TK1) has been studied extensively, primarily as a diagnostic biomarker for a variety of cancer types. TK1 plays a role in regulating the intracellular thymidine pools throughout the cell cycle, and thus regulates cell proliferation. [2] After cell division is completed, TK1 is degraded intracellularly, so that it does not pass to body fluids after normal cell division. However, this is not the case in cancer cells. In many cancer patients, serum TK1 level is significantly up-regulated. Higher serum TK1 activity levels correlate with a more advanced cancer stage and grade. [3] The mechanism of release of TK1 into the serum is poorly understood. My project aims to understand the cellular movement of TK1 at different cell cycle stages, and identify at what cell cycle stage TK1 is being released by cancer cells.
Methodology
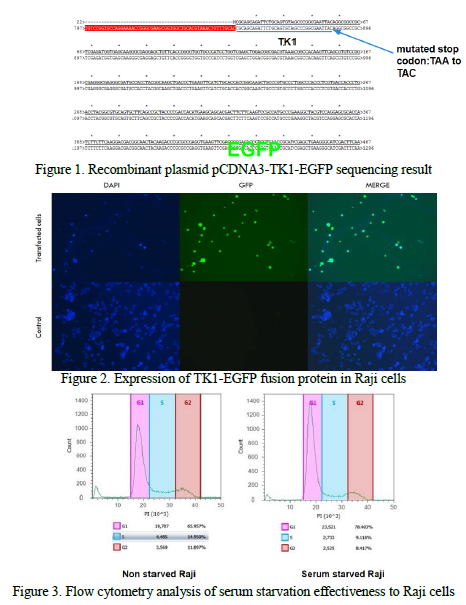
To trace the movement of TK1 during a cell cycle, TK1 gene was cloned into a bacterial plasmid vector pCDNA 3-EGFP. This recombinant plasmid pCDNA3-TK1-EGFP was transformed to E. Coli cells, and Ampicillin was used to select for single colonies. Recombinant plasmid was isolated from bacteria culture, and sequenced by BYU DNA Sequencing Center to make sure the TK1 gene and the GFP gene remain undisrupted. Afterwards, recombinant plasmid was transfected into rapidly dividing Burkitt’s lymphoma cells (Raji). Fluorescent microscopy was performed on transfected Raji cells to confirm the expression of TK1-EGFP fusion protein. Raji cells were serum starved to be synchronized into the same cell cycle stage for tracing fusion protein movement. Flow cytometry was performed to analyze serum starvation.
Results
Correct recombinant plasmid pCDNA3-TK1-EGFP was constructed and confirmed through DNA sequencing. Successful transfection of Raji cells was obtained and confirmed through fluorescent microscopy. Serum starvation of Raji cells was performed and analyzed by flow cytometry, but desired synchronization is still yet to be achieved and will need more
troubleshooting procedures.
Discussion
Through our studies, we have successfully obtained the fusion TK1-EGFP protein and can use it to trace TK1 movement during cell cycle stage. However, we do need to troubleshoot more on the synchronization procedure. Serum starvation is a traditional standard, but it didn’t work very well in our case. We have tried to find the optimal medium condition and starvation time, but haven’t found a good working protocol. Our future endeavors will be on looking for a different synchronization method for our cell cycle stage analysis.
Conclusion
TK1-EGFP expression can be used as an indicator of TK1 expression in mammalian cells. Serum starvation to obtain Raji cell synchronization is not a very successful method. More troubleshooting need to be performed to obtain a better synchronization protocol. Further research is needed for the next stage of this project.
Reference
1. American Cancer Society. Cancer Facts & Figures. Cancer Facts Fig. 2014.
2. Ji Zhou, Ellen He, Sven Skog. The proliferation marker thymidine kinase 1 in clinical use. Molecular and Clinical Oncology, vol. 1 iss:1 pg:18 -28, 2013.
3. Alegre MM, Weyant MJ, Bennett DT, Yu JA, Ramsden MK, Elnaggar A, Robison RA, O’Neill KL. Serum detection of thymidine kinase 1 as a means of early detection of lung cancer. Anticancer research, vol. 34 iss:5 pg:2145 -51, 2014.
