Tyler White and Bradford Berges, Microbiology and Molecular Biology
Introduction:
The bacterium Staphylococcus Aureus is the cause of a serious skin disease that is known to cause life-threatening infections such as pneumonia, endocarditis, meningitis, sepsis, bacteremia, and toxic shock syndrome [1]. Nonetheless, most SA infections are readily remedied via antibiotic treatment with methicillin. However, over the course of frequent exposure to various antibiotics, the bacteria have evolved a mutant strain (MRSA) that is completely resistant to the drug. In 2005 an estimated 11,000 deaths occurred that can be attributed to SA, with the majority caused by MRSA isolates [2]. Albeit the mutant is named MRSA, isolates of this strain are usually resistant to all known antibiotics with vancomycin being the common exception. Approximately half of all SA infections in the United States are resistant to penicillin, methicillin, tetracycline, and erythromycin [3]. The need for antibiotic alternative treatment for MRSA, i.e. phage therapy, is becoming increasingly crucial as there are few drugs left that the bacteria (such as vancomyocin) has yet to develop resistance towards; once MRSA has acquired such immunities, phage therapy may become the only feasible method of treatment for this bacterial infection. The primary mission of my project is to isolate numerous samples of phage from natural sources that are highly virulent and also have a broad tropism with respect to MRSA strains. We plan to aggregate these phage into a specialized cocktail used to treat and eradicate the bacterium MRSA in infected patients. Once isolated, these phage isolates will have their genomes sequenced (to determine novelty) and thereafter published.
Methodology:
LB agar plates were used to passage bacteriophage species against our library of MRSA strains. Low agarose top agar was used as medium through which phage amplification (via interaction with MRSA cells) took place and respective virulence of each phage species was determined. An ultracentrifugation procedure and a Norgen DNA Isolation kit were utilized to obtain phage DNA samples. Restriction enzyme digests were performed to cut DNA and thereafter ligation reactions were conducted to insert phage DNA into cloning vectors. Plasmid transformations were completed using E. coli bacterial cells. A Plasmid preparation kit was used to extract DNA from transformed cells and Nanodrop tests were run to determine concentration of DNA. Plasmid samples were finally submitted for cycle sequencing.
Results:
High titer lysates of the virulent M1CJ11 phage were produced efficiently, but extraction of phage DNA required much troubleshooting. An ultracentrifugation protocol was created and used to prepare a phage sample of inordinately high concentration so as to yield a successful DNA extraction product. Desired results for DNA digestion were confirmed via gel electrophoresis. Blue-white screening tests indicated that the transformation of phage DNA-containing plasmids into E. coli cells was completed successfully. Plasmid DNA was amplified and submitted for sequencing.
 Figure 1: Phage Ultracentrifucation
Figure 1: Phage Ultracentrifucation
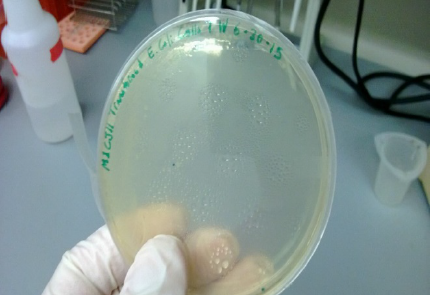 Figure 2: Phage Plasmid Transformation in E. Coli
Figure 2: Phage Plasmid Transformation in E. Coli
Discussion:
Initial samples were submitted and did not return a DNA sequence characteristic of any known phage species. This could possibly be attributed to an ineffective vector ligation reaction in which only a small proportion of phage inserts were able to be incorporated into their target plasmids. Secondary samples experienced sequencing errors that may have been caused by a concentration insufficiency. In the future CIP preparation will likely be implemented prior to ligation of vectors so as to ensure a higher ratio of plasmids (used for transformation) that have incorporated phage DNA.
Conclusion:
The bacteriophage species in our possession (such as M1CJ11) have been passaged against several MRSA strains to prove their level of virulence. We have also developed an effective (albeit tedious) process through which we can directly isolate purified genomic bacteriophage DNA. Phage plasmid amplification by means of E. coli bacterial cell transformation has been properly completed with confirming results. Therefore, the pathway has now been paved for the genomic sequencing of myriad phage species to take place in an expeditious fashion. Our aims for future research include sequencing and comparing genetic characteristics exhibited by species common to families of bacteriophage such as Myoviridae, Siphoviridae, and Podoviridae.
References:
- Lowy, F.D., Staphylococcus aureus infections. N Engl J Med, 1998. 339: p. 520-532.
- Klein, E., D.L. Smith, and R. Laxminarayan, Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis, 2007. 13: p. 1840-6.
- Albrich, W.C. and S. Harbarth, Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis, 2008. 8: p. 289-301.
