Amber Brown and Dr. Julianne Grose, Microbiology
Introduction
Cholera, the Disease: Cholera is a disease that affects three to five million people each year with approximately 100,000 deaths. Transmitted mainly by the drinking-‐water supply, it causes an infection in the small intestine leading to severe diarrhea and vomiting. If left untreated, it can cause death within hours. The disease is caused by the bacterium Vibrio cholerae which was first isolated by Filippo Pacini in 1854. Vibrio cholerae is gram negative, comma shaped, and flagellated. It is easily treated with modern water purification techniques, and as such it is most prevalent in developing countries that lack this infrastructure. Countries that are recovering from natural disasters are especially vulnerable. One of the first documented outbreaks occurred in 1817 near the Ganges River. British trade ships unknowingly spread the disease by transporting infected bilgewater from the Bay of Bengal. Several major outbreaks followed in places including as London, New York, and several parts of Russia. What is needed is a safe, cheap way to detect and destroy cholera in water. Our solution: using the natural mechanisms of quorum sensing, phage, and biofilm inhibitors to purify contaminated water sources.
Quorum Sensing: Quorum sensing is the cell-‐to-‐cell signaling system that allows for coordinated behavior between bacteria through the secretion and detection of signaling molecules termed autoinducers. Some autoinducers are specific to a certain bacteria, while others are detectable by a broad range of bacteria. The N-‐acyl-‐L-‐homoserine lactones (AHLs) are a class of autoinducer secreted by many Gram-‐negative bacteria, including Vibrio cholerae. While Escherichia coli does not produce AHLs, it does have the SdiA receptor necessary to detect these autoinducers. The binding of AHLs from V. cholerae to the SdiA receptor on E. coli triggers a signal cascade that activates the transcription of quorum sensing linked genes in E. coli. By inserting our target genes into the quorum sensing linked section of E. coli’s DNA, we can design a specific response to the detection of V. cholerae by E. coli. The specific response we are trying to achieve is induction of the lytic cycle of prophage lambda in E. coli. We hypothesize that SdiA, a transcriptional activator in E.Coli, senses cholera’s autoinducer molecules. To test this, we have infected SdiA wild type and SdiA-‐knockout E.coli with bacteriophage lambda, and have performed top agar plaque-‐assay tests to confirm that lambda induction is SdiA-‐dependent. (Figure 2 in Results).
Biofilm: Bacteria living in a biofilm are physiologically different from a bacterium of the same species in a planktonic state. In a biofilm, cells are fixed within a self-‐produced extracellular polymeric substance (EPS.) Vibrio cholerae‘s virulency is one of the physiological changes it undergoes when changing its physical sate. The formation of its biofilm is reciprocally controlled by two chemical signaling systems: 3’,5’-‐cyclic diguanylic acid (c-‐di-‐GMP,) which activates biofilm formation, and quorum sensing, which represses it. Many species of bacteria in the Vibrio genus form biofilms, although they are very different in their structure and stability. Vibrio cholerae produces a biofilm that is free floating rather than one that attaches to solid surfaces which presents major issues in characterizing and running degradation assays on it. We will try to disrupt and degrade the protective biofilm of cholera by using alpha-‐Amylase and Dispersin B. These are known biofilm inhibitors. The goal is to attach one or both of these inhibitors to the tail fibers of the lambda phage that will be released from E. coli, so that when E. coli senses cholera nearby, it will release the phage, killing the E. coli and attacking the biofilm of cholera, effectively killing the organism and purifying the water source.
Methodology
All standard aseptic techniques and practices were used in this study. V. cholerae was plated on LB plates for all experiments. We were able to create an LB and sea salt medium, which allowed cholera to form a biofilm, something that had never been done before in a lab. Using this technique, we were able to run degredation assays on it and determine how effective our system was. The enzymatic activity of α-Amylase was characterized to determine its capacity to inhibit biofilm formation by V. cholerae. Samples were prepared by adding 50 μL of V. cholerae culture to 1 mL of a high salt LB. The samples were then treated by the addition of purified α-Amylase in various concentrations and allowed to incubate at 30°C for 48 hours. After 48 hours the samples were examined and a distinct difference was seen in the amount of biofilm formed in the treated and untreated samples (images not shown). The samples were then transferred to eppendorf tubes and centrifuged at 16,000 × g for two minutes, the supernatant was discarded to remove the growth media, and the samples were resuspended in 200 μL ddH2O. The samples were then stained with 50 μL of a 0.03% CV solution and allowed to incubate for five minutes. The samples were again centrifuged at 16,000 × g for two minutes and the supernatant 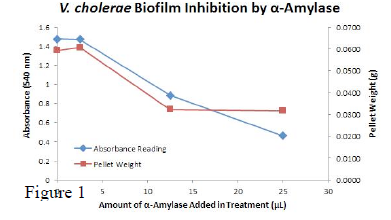 containing excess CV was discarded. The pelleted biofilm was then washed with 800 μL of 95% EtOH without resuspension, centrifuged for 30 seconds, and the EtOH was discarded. The EtOH wash was repeated twice more for a total of three washes. The samples were then resuspended in 200 μL EtOH, transferred to a 96-well plate, and incubated for five minutes. The plate was then shaken for ten seconds and absorbance readings were taken for each sample at 540 nm, the absorbance wavelength of CV. The graph to the left (Figure 1) shows both the average pellet weight and the average absorbance readings for samples treated with 0, 2.5, 12.5, and 25 μL of α-Amylase. There is a distinct reduction in the amount of biofilm growth between untreated and treated samples, with samples treated with 25 μL α-Amylase showing a 65.8% decrease in biofilm formation after 48 hours. While this clearly shows the ability of α-Amylase to inhibit biofilm formation by V. cholerae, further characterization is needed to determine the capacity of α-Amylase to degrade preexisting biofilms. The biofilm assay was again run with Dispersin B as the treatment enzyme, and similar data was gathered.
containing excess CV was discarded. The pelleted biofilm was then washed with 800 μL of 95% EtOH without resuspension, centrifuged for 30 seconds, and the EtOH was discarded. The EtOH wash was repeated twice more for a total of three washes. The samples were then resuspended in 200 μL EtOH, transferred to a 96-well plate, and incubated for five minutes. The plate was then shaken for ten seconds and absorbance readings were taken for each sample at 540 nm, the absorbance wavelength of CV. The graph to the left (Figure 1) shows both the average pellet weight and the average absorbance readings for samples treated with 0, 2.5, 12.5, and 25 μL of α-Amylase. There is a distinct reduction in the amount of biofilm growth between untreated and treated samples, with samples treated with 25 μL α-Amylase showing a 65.8% decrease in biofilm formation after 48 hours. While this clearly shows the ability of α-Amylase to inhibit biofilm formation by V. cholerae, further characterization is needed to determine the capacity of α-Amylase to degrade preexisting biofilms. The biofilm assay was again run with Dispersin B as the treatment enzyme, and similar data was gathered.
Results
 We were able to see the effects of plating cholera on E. coli with a lambda prophage inside of it (top plate, Figure 1) We discovered that lysogenic bacteriophage lambda responds to V. cholera by lysing its host. The bottom plate in Fig. 1 is E. coli with no cholera. We believed it was the SdiA receptor detecting quorum sensing molecules from cholera that was inducing lysis, though tests that we ran using GFP showed that SdiA was not specific for cholera. Our next step was to mutate SdiA, which we did using selective promotes. We were able to change some of the codons in SdiA to look more like CqsS, the CAI-‐1, or quorum sensing binding protein. This way, mutated SdiA would bind only cholera-‐specific lytic inducing molecules. After inducing lysis, the phage would need to have biofilm inhibitors attached to their tail fibers to destroy cholera in the water. Two biofilm inhibitors were tested, and both inhibited the formation of biofilm by 40% or more, though we still need to test on existing biofilms.
We were able to see the effects of plating cholera on E. coli with a lambda prophage inside of it (top plate, Figure 1) We discovered that lysogenic bacteriophage lambda responds to V. cholera by lysing its host. The bottom plate in Fig. 1 is E. coli with no cholera. We believed it was the SdiA receptor detecting quorum sensing molecules from cholera that was inducing lysis, though tests that we ran using GFP showed that SdiA was not specific for cholera. Our next step was to mutate SdiA, which we did using selective promotes. We were able to change some of the codons in SdiA to look more like CqsS, the CAI-‐1, or quorum sensing binding protein. This way, mutated SdiA would bind only cholera-‐specific lytic inducing molecules. After inducing lysis, the phage would need to have biofilm inhibitors attached to their tail fibers to destroy cholera in the water. Two biofilm inhibitors were tested, and both inhibited the formation of biofilm by 40% or more, though we still need to test on existing biofilms.
Conclusion
While we did not unite all of the pieces to the complex puzzle of creating a safe, cheap, and easy way to eradicate cholera from water sources, we have solved many of the preliminary steps. We were able to make SdiA specific for E. coli, prove that cholera does induce the lytic lifecycle of prophage lambda, and innovate a way to grow cholera biofilm in the lab, allowing us to test different biofilm disrupters. These were all novel and important discoveries. For future directions, we could investigate the effects of other prophage, and new biofilm disrupters that will not harm the environment. Furthermore, we are researching an alternative method for lambda induction, utilizing the cro regulation system that lambda uses. There is plenty of work still to be done, and hopefully our positive results will continue.
