Sudeep Ghimire and Dr. Craig Coleman, Department of Plant and Wildlife Sciences
Bromus tectorum, commonly called cheatgrass or downy brome is an exotic annual invasive weed which was introduced to the western United States over hundred years ago. B. tectorum has quickly expanded its range, displacing native flora and causing extensive loss of shrub and rangeland habitats along with supporting devastating wildfires, especially in the intermountain west region. Consequently, this invasive weed’s control is a growing concern. However, current control methods being used like control burning, herbicides, etc. are non-specific and mostly ineffective. Methods of biological control are currently under investigation, of which one of them is using the fungus, Pyrenophora semeniperda (aka Black Fingers of Death, BFOD). Because cheatgrass genotypes exhibit varying levels of resistance to BFOD and other methods of biological control, it would be helpful to know which genotypes are present in which geographical areas. In order to develop effective control strategies, genotyping individuals from different populations of cheatgrass are required.
SNPs are robust markers used to distinguish between different individual genotypes. SNP markers will allow us to understand the genotypic diversity and distribution of cheat grass and enact control strategies. So, the whole goal behind the project was to develop certain number of SNPs and genotype various cheatgrass populations and compare the SNP genotyping with the genotyping data done using the SSR and allozyme markers. As previous studies have used allozyme or microsatellite markers, both of which demonstrate high homozygosity which might be due to the use of no more than 5 markers to genotype individual plants, thus underrepresenting the actual heterozygosity. However, after the SNP development and genotyping process, we were able to find few heterozygous lines which even seemed to be less accountable. In order to be more ascertained about the outcrossing level and heterozygousity level, we have been working on two other projects namely cheatgrass outcrossing study and heterozygousity study. The results from these projects will allow us to give more clues regarding the heterozygosity outcrossing levels in cheatgrass.
In order to test my hypothesis, we developed 99 SNPs out of the total of 243 SNPs that we got from Primer Picker software with a validation rate of nearly 45%. Over 22,000 putative SNPs that were identified previously through 454 sequencing of cheatgrass cDNA, from which those 243 SNPs were chosen for validation. After designing the primer pairs and analyzing using the KASP SNP validation from KBioSciences, 23 different lines of cheatgrass individuals were validated. Then 288 individuals that represent the most abundant SSR genotypes from 95 different populations from western US were genotyped using the validated SNPs.
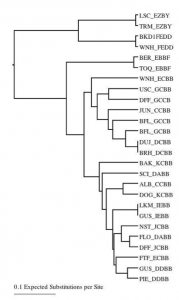
As the preliminary evaluation, we found out that some of the warm desert habituated cheatgrass individuals such as EZBY and FEDD show huge difference from rest of the others in the phenotypic tree as shown in the figure 0.1. However, EBBF which is more similar to the warm desert cheatgrass lines grouped with other normal lines as ECBB, GCBB, etc. We can be clearer when we will find out how they will behave in the outcrossing study.
Further, we chose 96 validated SNPs using the Fluidigm protocol genotyped 96 different lines getting around 10000 data in a single run. We plan on modifying the Fluidigm SNP genotyping protocol using 24 SNPs which will allow us to genotype 384 individuals in a single run and this will be far more economical and convenient. To conclude, we were able to develop successful rate of validated SNPs and genotype various population of cheatgrass individuals, thus relating various microsatellite and allozyme data. This project allowed me to develop my capability of working in the lab in more efficient way.
References
- D. Chagne et al. (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353-358.
- Ramakrishnan A.P., Coleman C.E., Meyer S.E., Fairbanks D.J. (2002) Microsatellite markers for Bromus tectorum (cheatgrass). Molecular Ecology Notes 2, 22-23.
- Ramakrishnan A.P., Meyer S.E., Waters J., et al. (2004) Correlation between molecular markers and adaptively significant genetic variation in Bromus tectorum (Poaceae) an inbreeding annual grass. AmericanJjournal of Botany 91, 797-803.
- Ramakrishnan A.P., Meyer S.E., Fairbanks D.J., & Coleman C.E. (2006) Ecological significance of microsatellite variation in western North American populations of Bromus tectorum. Plant Species Biology 21:61-73